Signalling pathways and causal depth
Generating deep signalling pathways with refinements
Yvan Sraka & Younesse Kaddar
I. Biological context
II. Theoretical Model
III. Refinements
IV. Implementation
I. Biological context
Why does evolution favor deep signalling pathways?
I. Biological context
Why does evolution favor deep signalling pathways?
Why does evolution favor deep signalling pathways?
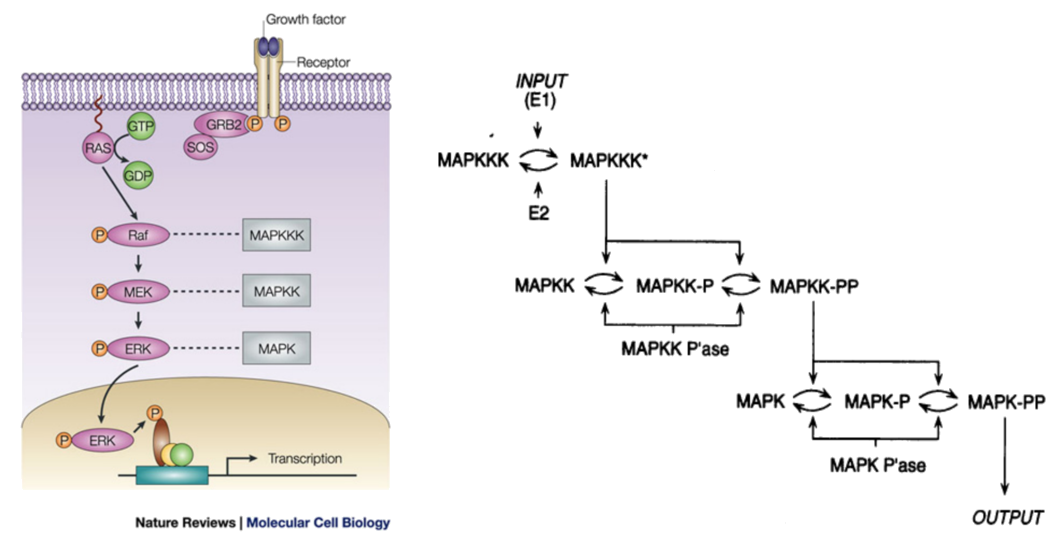
Example: the MAPK/ERK pathway
Animation courtsey of Ribosome Studio Youtube channel
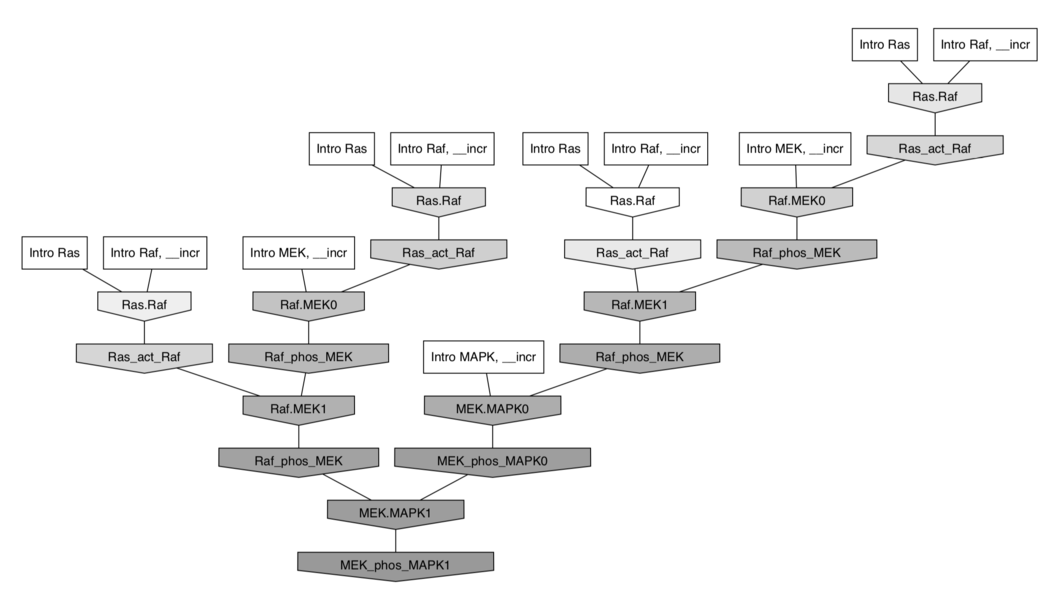
$⇓ \;$ in Kappa
Why does evolution favor deep signalling pathways?
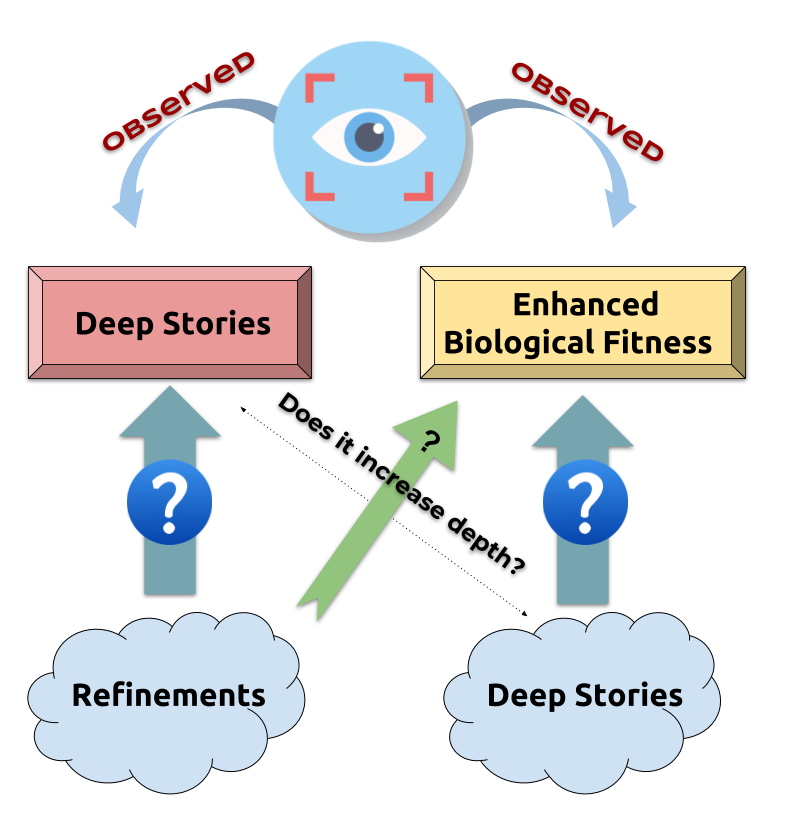
Natural selection: Deeper stories $⟹$ Improving biological fitness
BUT counter-intuitive
-
energetically
-
probabilistically: Indeed: if
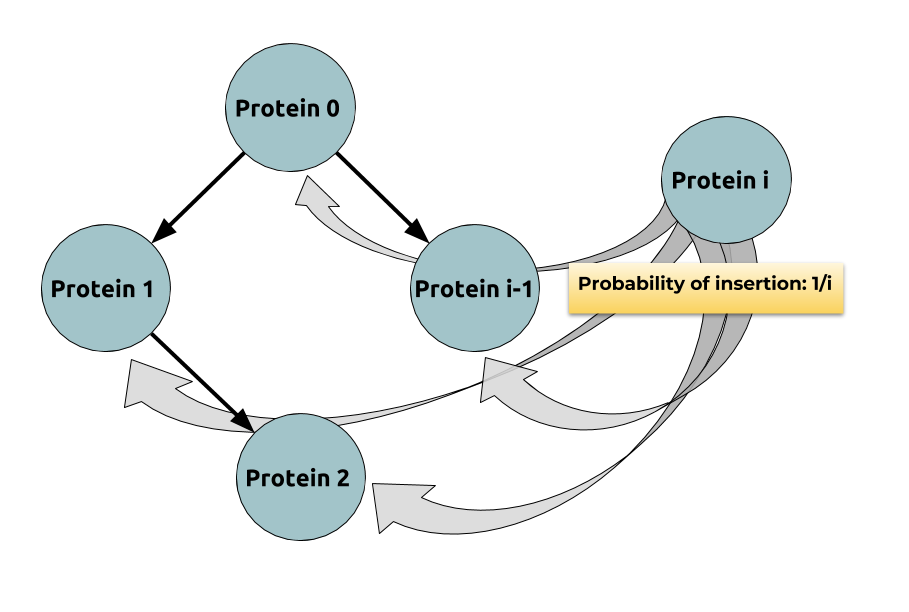 $$ \begin{align*} & 𝔼(\max_i \depth(P_i)) \\ & ≤ 𝔼\Big(\ln\Big(\underbrace{\sum\limits_{ i=0 }^n \exp(\depth(P_i))}_{\; ≝ \; α_n}\Big)\Big) \\ & ≤ \ln \underbrace{𝔼(α_n)}_{\rlap{\substack{= \; 𝔼(α_{n-1}) + 𝔼(\exp(\depth(P_n))) \\ = \, (1 + \frac e i) \, 𝔼(α_{n-1})}}}\\ & = \ln \Big(\prod\limits_{ i=1 }^n \Big(\underbrace{1 + \frac e i}_{≤\, \exp (\frac e i)}\Big) \Big) = e H_n \sim \boxed{e \ln n} \end{align*} $$
$$ \begin{align*} & 𝔼(\max_i \depth(P_i)) \\ & ≤ 𝔼\Big(\ln\Big(\underbrace{\sum\limits_{ i=0 }^n \exp(\depth(P_i))}_{\; ≝ \; α_n}\Big)\Big) \\ & ≤ \ln \underbrace{𝔼(α_n)}_{\rlap{\substack{= \; 𝔼(α_{n-1}) + 𝔼(\exp(\depth(P_n))) \\ = \, (1 + \frac e i) \, 𝔼(α_{n-1})}}}\\ & = \ln \Big(\prod\limits_{ i=1 }^n \Big(\underbrace{1 + \frac e i}_{≤\, \exp (\frac e i)}\Big) \Big) = e H_n \sim \boxed{e \ln n} \end{align*} $$
Possible explanations:
- Greater depth:
- $$\, \\ \\ \\ \begin{cases} \text{ enhances stability } \\ \text{ deletions make pathways non-functional more often than additions do } \end{cases}$$
II. Theoretical model
Does refinement increase depth?
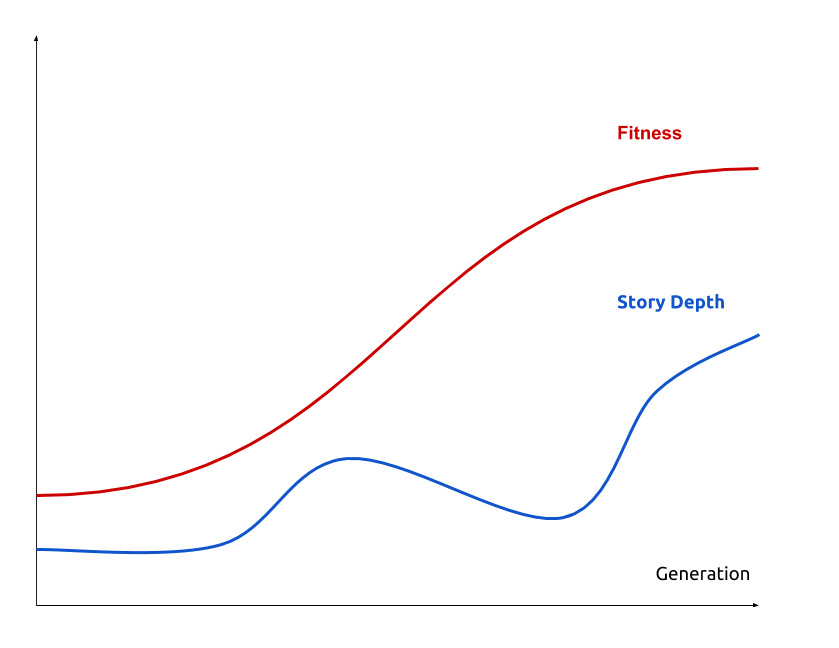
Genetic algorithm
An heuristic optimisation that loop on two steps:
- MUTATION: map ancesters to $n$ descendent with random mutations
- SELECTION: filter descendent on their fitness (ERK quantity)
Model biais: biological clock assumption, spontaneous discrete generation, etc …
III. Refinements
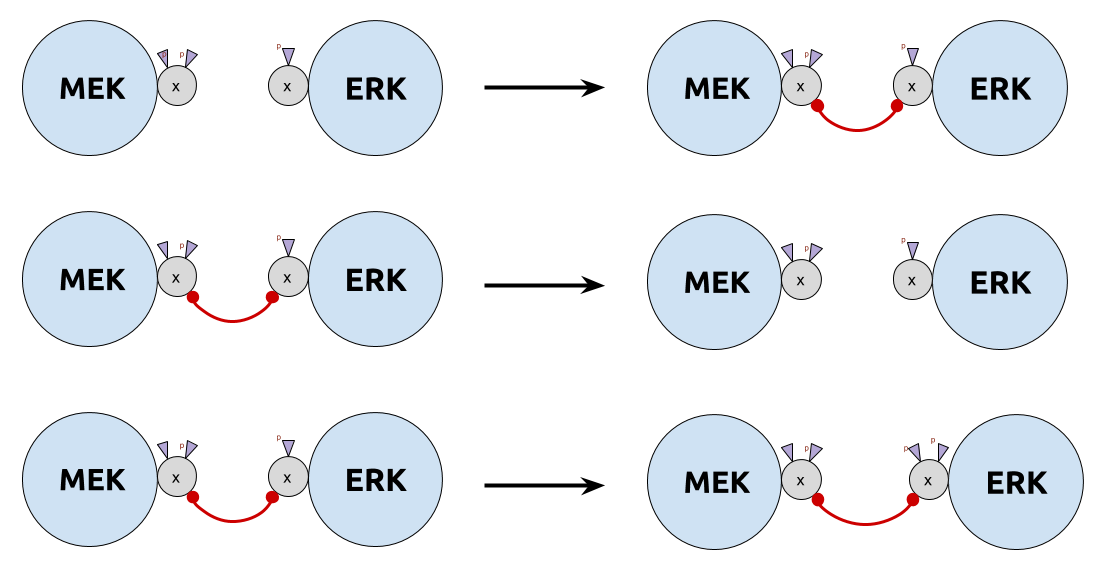
- $\color{DarkCyan}{τ_u}$ too big: unbind before phosphorylation (too liquid)
- $\color{DarkCyan}{τ_u}$ too small: never unbind (too sticky)
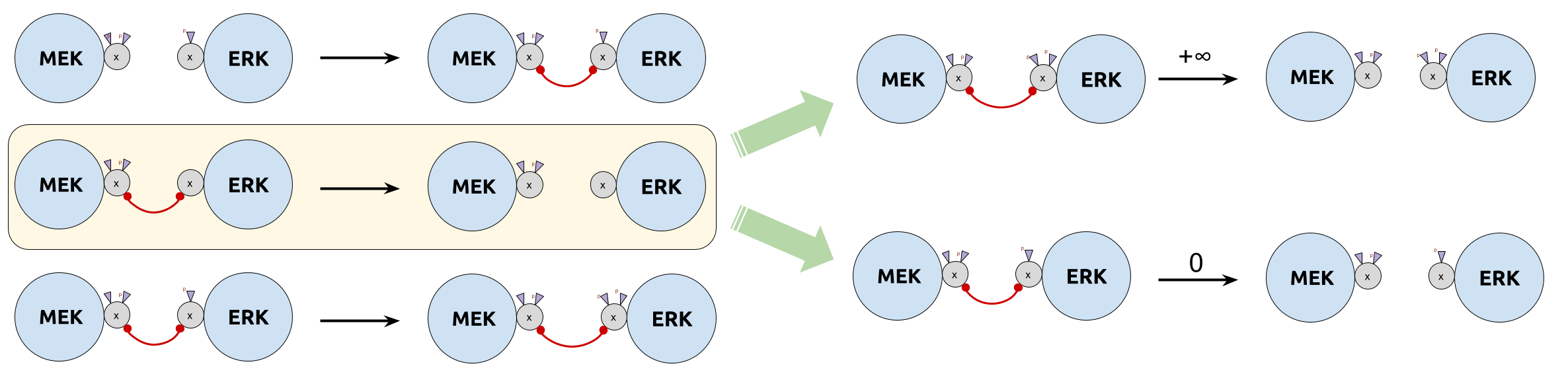
Category of site graphs: $\SGph$
- Objects $(V, λ, σ, μ)$:
-
- $V$: set of nodes
- $λ: V ⟶ 𝒜$: name assignment
- $σ: V ⟶ 𝒫(𝒮)$: site assignment
- $μ: \underbrace{\text{ matching}}_{\rlap{\text{irreflexive symmetric binary relation}}}$ over $\sum\limits_{ v ∈ V } σ(v)$
- Morphism $f: (V, λ, σ, μ) ⟶ (V', λ', σ', μ')$:
- Name/site/edge-preserving and edge reflecting monomorphism $f: V ⟶ V'$
- Signature $Σ: 𝒜 ⟶ 𝒮$:
-
- $x ≤ Σ \quad ⟺ \quad σ_x(V_x) ⊆ Σ(λ_x(V_x))$
- $Σ ≤ x \quad ⟺ \quad Σ(λ_x(V_x)) ⊆ σ_x(V_x)$
Epimorphisms
- Forgetful functor to the category of graphs:
- $$U: \SGph ⟶ \Gph$$
- Epimorphisms from $x$ to $y$:
- $$[x, y]^e \; ≝ \; \big\lbrace h ∈ \underbrace{\Hom[\SGph]{x, y}}_{\text{denoted by } [x,y]} \; \mid \; h \text{ is an epi (i.e. right-cancellable)} \big\rbrace$$
Lemma:
$$h ∈ \overbrace{\Hom[\SGph]{x, y}}^{\text{denoted by } [x,y]} \text{ is an epi } \\ \; ⟺ \; ∀ c_y ⊆ y \text{ connected component}, \, h^{-1}(c_y) ≠ ∅$$
Epi-mono factorisation
- Factorisation:
- $f ∈ [s, x]$ is said to be factored by $t$ if $f = γ ϕ$ for $ϕ ∈ [s, t]^e$ and $γ ∈ [t, x]$
Every iso $α ∈ [t, t']$ conjugates the factorisations $ϕ, γ$ and $ϕ', γ'$:
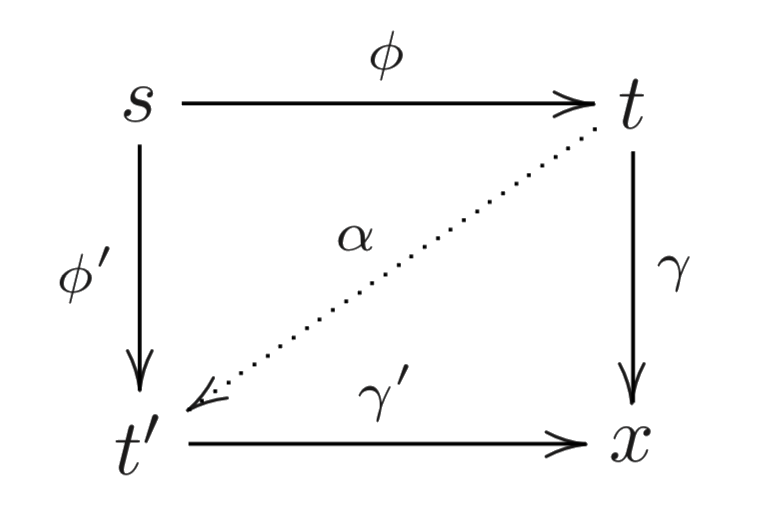
⟹ Equivalence relation:
The group $[t, t]$ acts freely on $[s, t]^e × [t, x]$ (as we have epis and monos), so by Burnside’s lemma:
Object refinement
- A refinement $Σ(s)$ of $s ≤ Σ$ under $Σ$:
- is a set comprised of one element per isomorphism class in $\lbrace t : Σ \; \mid \; [s, t]^e ≠ ∅ \rbrace$
Theorem: if $s ≤ Σ$ and $x : Σ$:
$$[s,x] \; ≅ \; \sum\limits_{ t ∈ Σ(s) } [s, t]^e ×_{[t,t]} [t, x]$$
Rule refinement
- Well defined atomic action (in a rewriting rule):
-
edge addition/deletion (when permitted), agent addition/deletion
- Labelled transition:
-
$$x \quad ⟶_f^R \quad f(α) \cdot x$$
where
- the rule $R \; ≝ \; \underbrace{s}_{\text{object}}, \underbrace{α}_{\text{action}}, \underbrace{τ}_{\text{rate}}$, whose activity $a(x, r) \; ≝ \; τ \vert [s, x] \vert$
- $f ∈ [s, x]$
If $θ$ is an iso: $θ(r) \; ≝ \; θ(s), θ(α), τ$ satisfies:
Rule refinement:
If $s ≤ Σ$, the refinement of the rule $R \; ≝ \; s, α, τ$ under $Σ$ is:$$Σ(s, α, τ) \; ≝ \; (t, ϕ(α), τ)_{t ∈ Σ(s), ϕ ∈ [s,t]^e/[t,t]}$$
IV. Methods
Our naive code implementation was split between:
- KaSimir: Genetic algorithm implementation, using KaSim to compute fitness
- KaStorama: wrapper around KaStor to retreive causal story depth from KaSim traces
- KaRapuce: Slitghly change rates of refined rules as mutation event
https://github.com/yvan-sraka/KaSimir
Demo time!
Bibliography
-
P. Boutillier, J. Feret, J. Krivine, and W. Fontana, “The Kappa Language
and Kappa Tools,” p. 52. -
“Signaling Pathways,” Tocris Bioscience. https://www.tocris.com/signaling-pathways.
-
V. Danos, J. Feret, W. Fontana, R. Harmer, and J. Krivine, “Rule-Based
Modelling, Symmetries, Refinements,” in Formal Methods in Systems
Biology, vol. 5054, J. Fisher, Ed. Berlin, Heidelberg: Springer Berlin
Heidelberg, 2008, pp. 103–122. -
E. Murphy, V. Danos, J. Féret, J. Krivine, and R. Harmer, “Rule-Based
Modeling and Model Refinement,” in Elements of Computational Systems
Biology, H. M. Lodhi and S. H. Muggleton, Eds. Hoboken, NJ, USA: John
Wiley & Sons, Inc., 2010, pp. 83–114. -
“List of signalling pathways,” Wikipedia. 30-Nov-2016.